NeuralPlexer
NeuralPlexer is a deep learning model designed for compound-protein docking. It combines graph neural networks (GNN) and self-attention mechanisms to accurately predict the structure of protein complexes and efficiently simulate the binding modes between compounds and protein targets. During the compound-protein docking process, NeuralPlexer provides confidence scores to assess the reliability of the predicted results, with scores ranging from 0 to 1. The closer the score is to 1, the higher the confidence in the docking result.
The advantage of NeuralPlexer lies in its ability to rapidly identify key binding sites, predict interactions between compounds and proteins, and optimize the accuracy of the docking structure. The model has broad applications in drug design, target discovery, and protein function analysis, providing high-precision computational support for drug screening and greatly improving research and development efficiency.
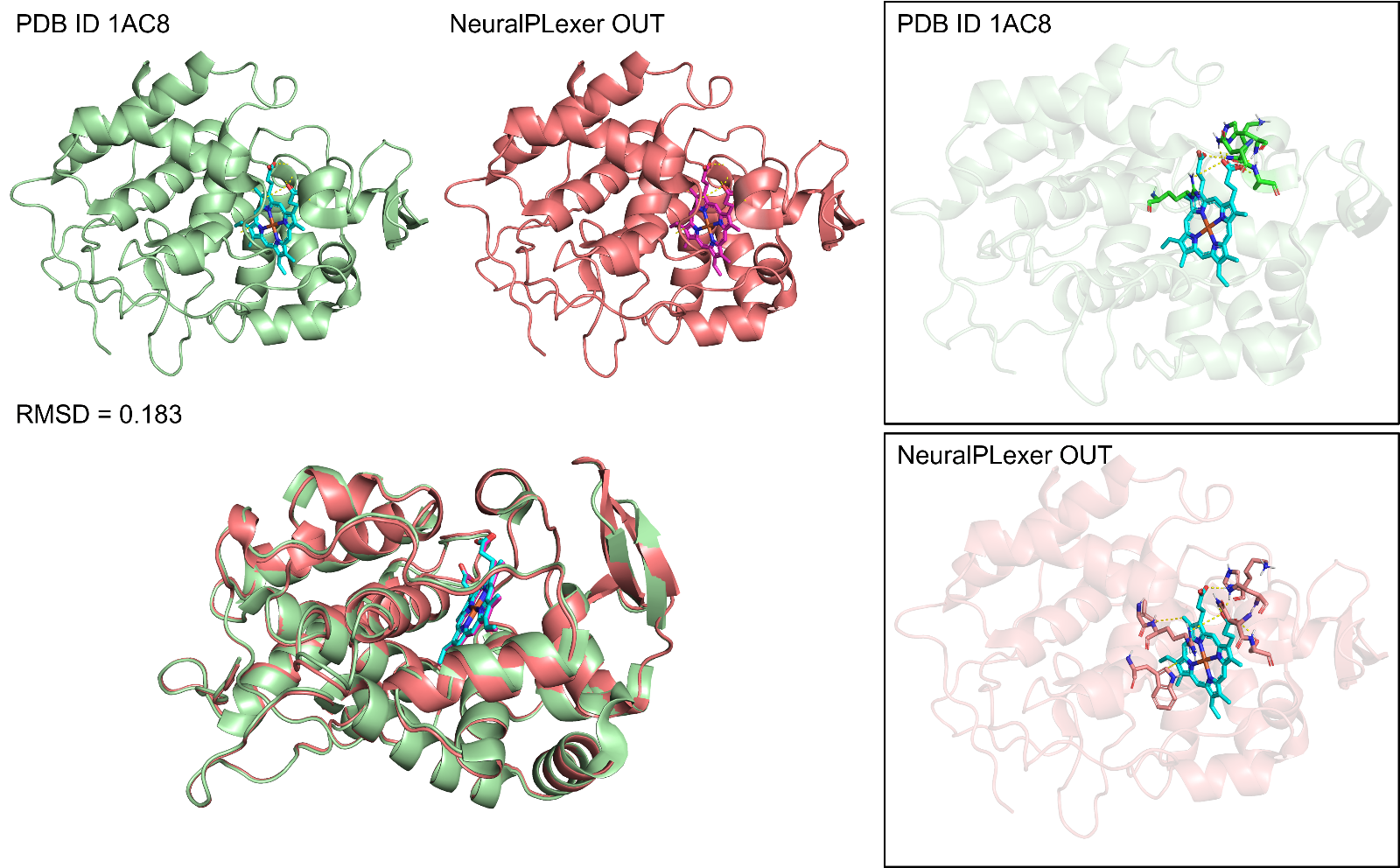
Figure 1: NeuralPlexer prediction of the compound-protein binding conformation, showing no significant difference from the experimentally resolved conformation (RMSD=0.183)