RoseTAAFold NA
RoseTTAFold NA is a high-precision deep learning tool specifically designed for nucleic acid and nucleic acid-protein complex modeling, expanding the application capabilities of the RoseTTAFold system in the field of nucleic acid structure prediction. This software efficiently performs nucleic acid monomer modeling, accurately predicts the three-dimensional structures of RNA and DNA molecules, and provides reliable solutions for nucleic acid-protein complex modeling.
RoseTTAFold NA integrates advanced multiple sequence alignment and deep learning algorithms to capture the key interactions between nucleic acid molecules and proteins, enabling the generation of precise complex structures. This modeling capability is crucial for studying the functions of nucleic acids, nucleic acid-protein binding mechanisms, and designing nucleic acid-based drugs. With its comprehensive support for nucleic acids and nucleic acid-protein complexes, RoseTTAFold NA is widely used in nucleic acid structure analysis, gene regulation research, and other fields, significantly accelerating the development of nucleic acid-related structural biology research.
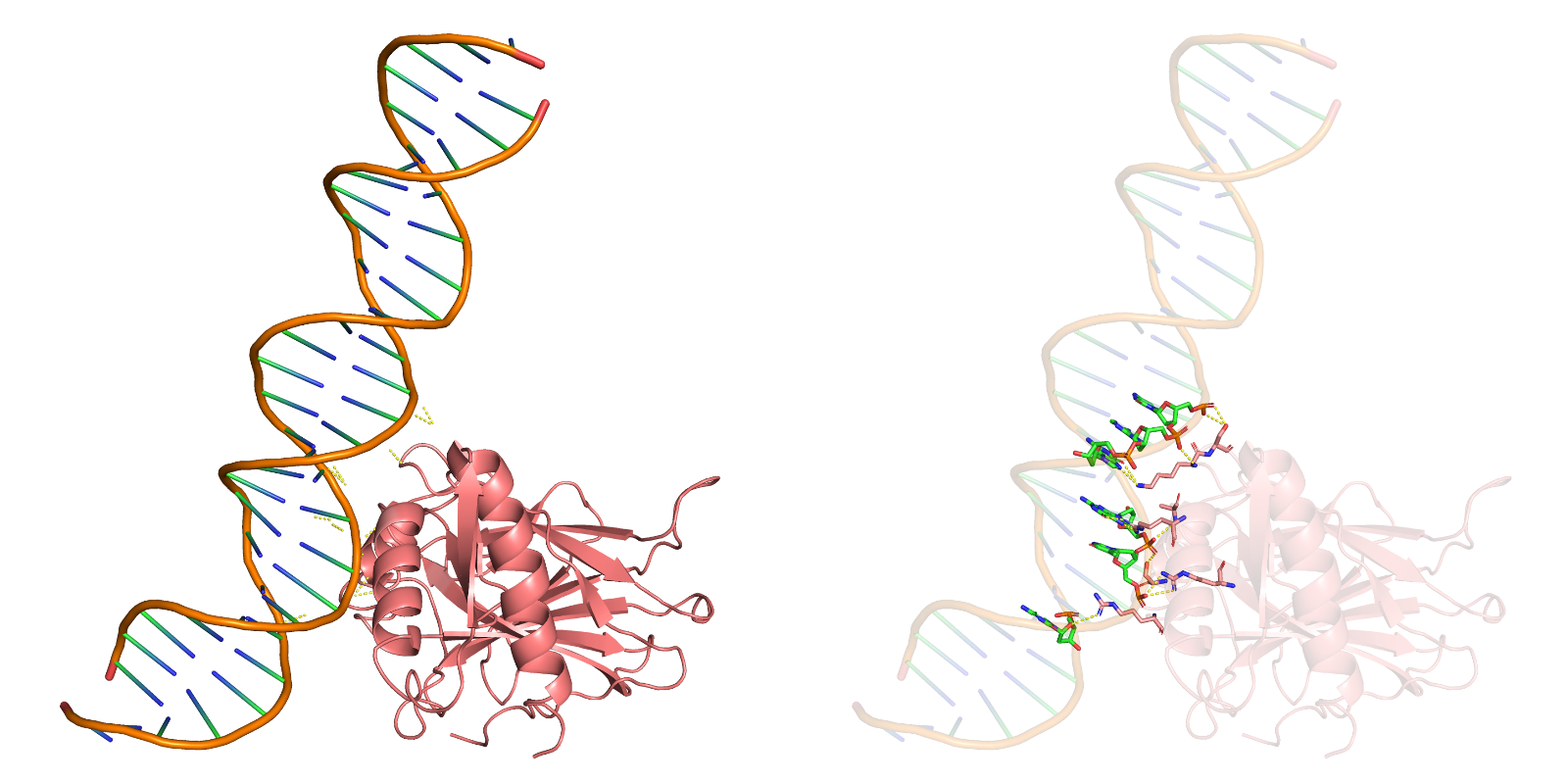
Figure 1: RoseTTAFold NA predicting the 3D model of DNA-protein complex.
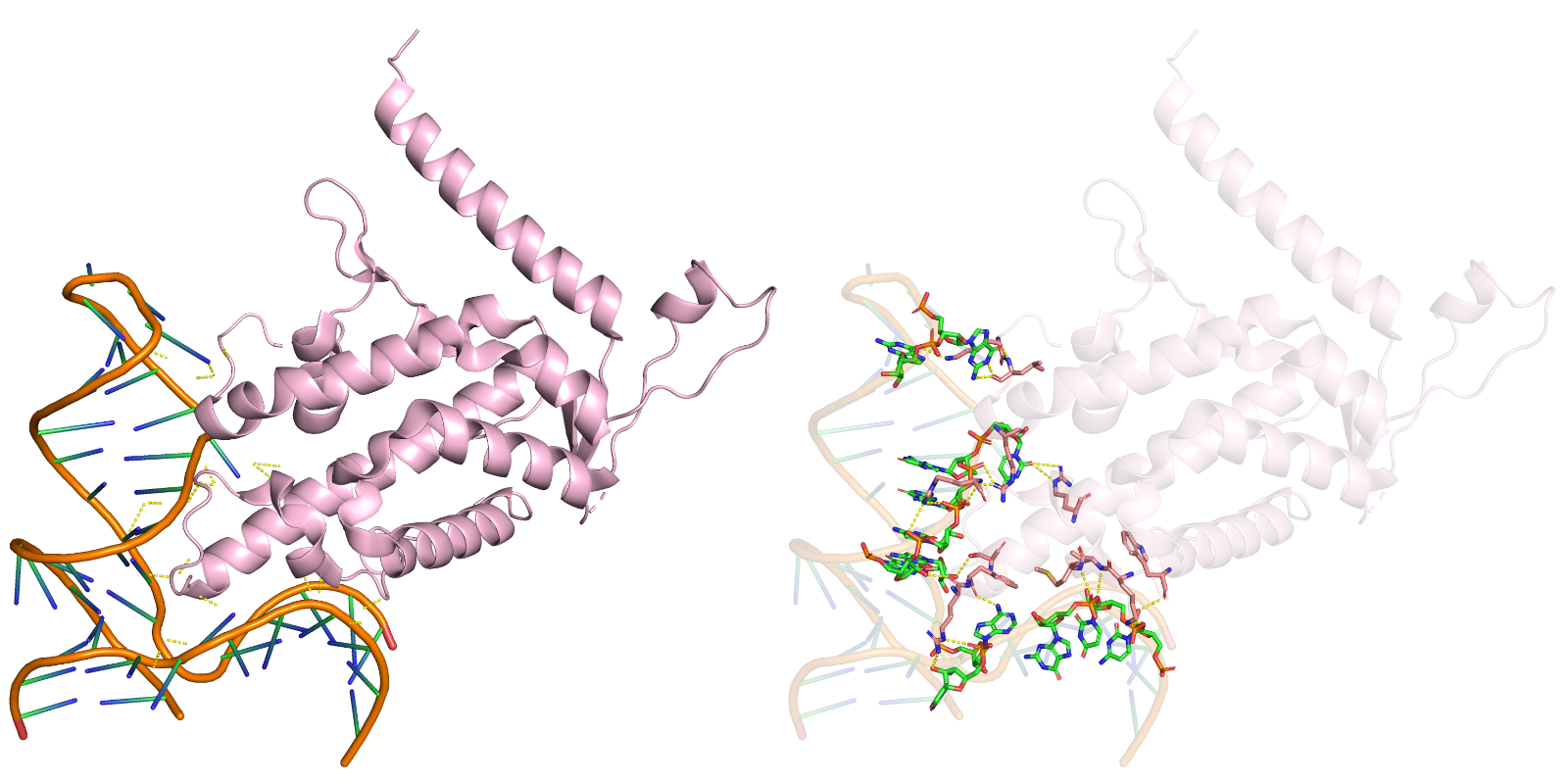
Figure 2: RoseTTAFold NA predicting the 3D model of RNA-protein complex.