OmegaFold
OmegaFold is an efficient and accurate protein structure prediction model specifically designed to address complex protein modeling tasks. It excels in both accuracy and efficiency, making it a crucial tool in the field of structural biology. Unlike traditional methods, OmegaFold can rapidly predict the three-dimensional structure of a protein directly from its sequence, without requiring a large amount of homologous template data, demonstrating its strong generalization capability.
The modeling accuracy of OmegaFold is attributed to its advanced deep learning architecture, which combines multimodal data training and optimization algorithms. This allows it to capture implicit information within the protein sequence and accurately predict key structural features. Its predictions achieve high resolution even in the absence of homologous templates or when dealing with low-homology sequences, particularly excelling in the modeling of complex domains and large-scale protein complexes.
In terms of efficiency, OmegaFold uses an optimized computational framework, significantly reducing prediction time. Compared to traditional physical modeling or experimental methods, OmegaFold has a clear advantage in handling large-scale sequence modeling tasks, enabling the rapid prediction of vast numbers of protein sequences, thus supporting high-throughput structural biology research.
OmegaFold has a broad range of applications, spanning basic research, protein engineering, and more. With its high precision and efficiency in modeling, researchers can quickly obtain the three-dimensional structures of proteins, providing important tools for exploring protein functions, designing novel proteins, and developing targeted drugs. With its outstanding performance, OmegaFold has established a pivotal position in the field of protein structure prediction, becoming a powerful tool driving advances in life sciences research.
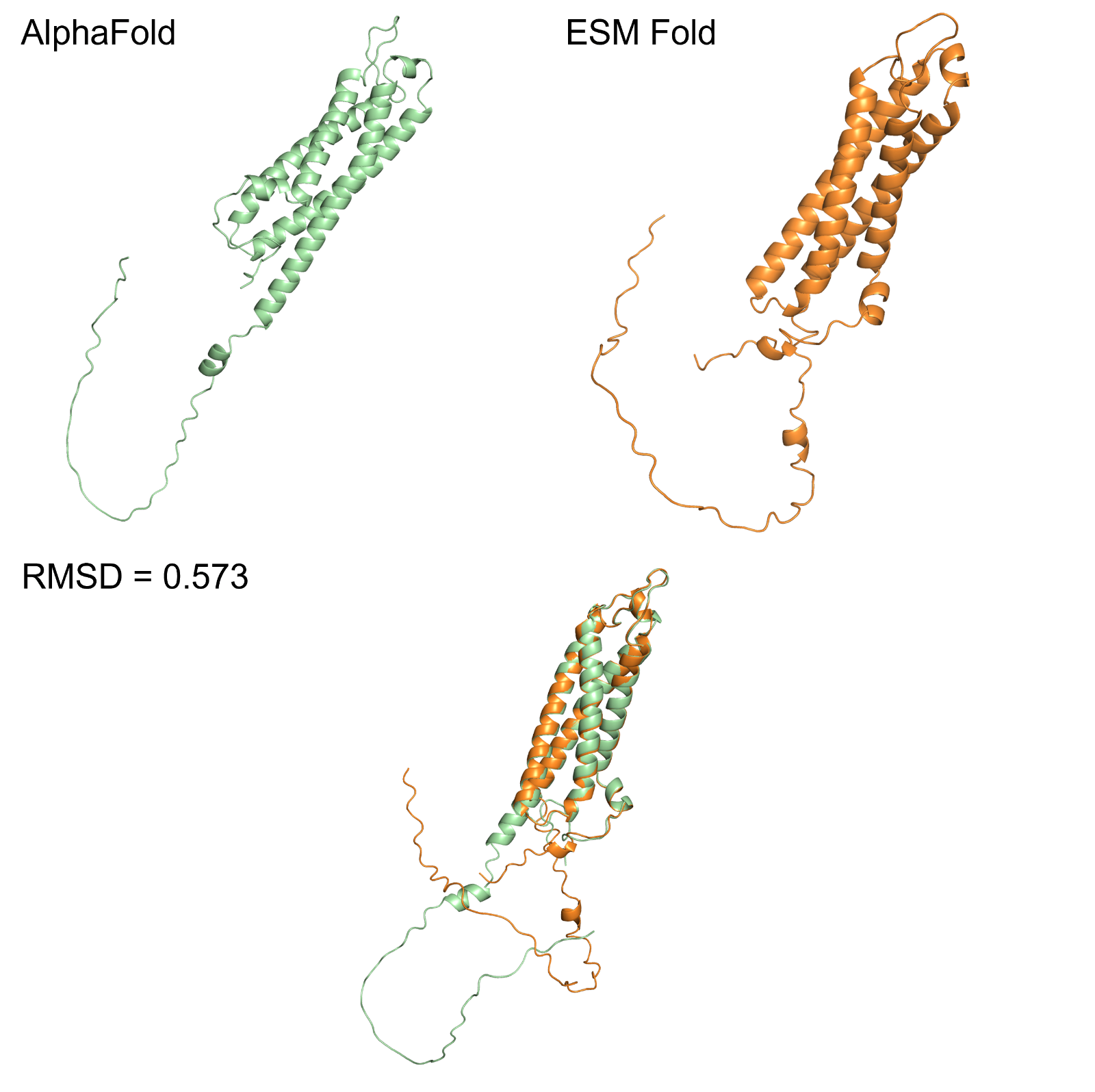
Figure 1: OmegaFold prediction of the protein model, showing minimal difference from the AlphaFold prediction model (RMSD=0.573).